Jian Hu, Xiangjie Li, Gang Hu, Yafei Lyu, Katalin Susztak, Mingyao Li
Published in Nature Machine Intelligence
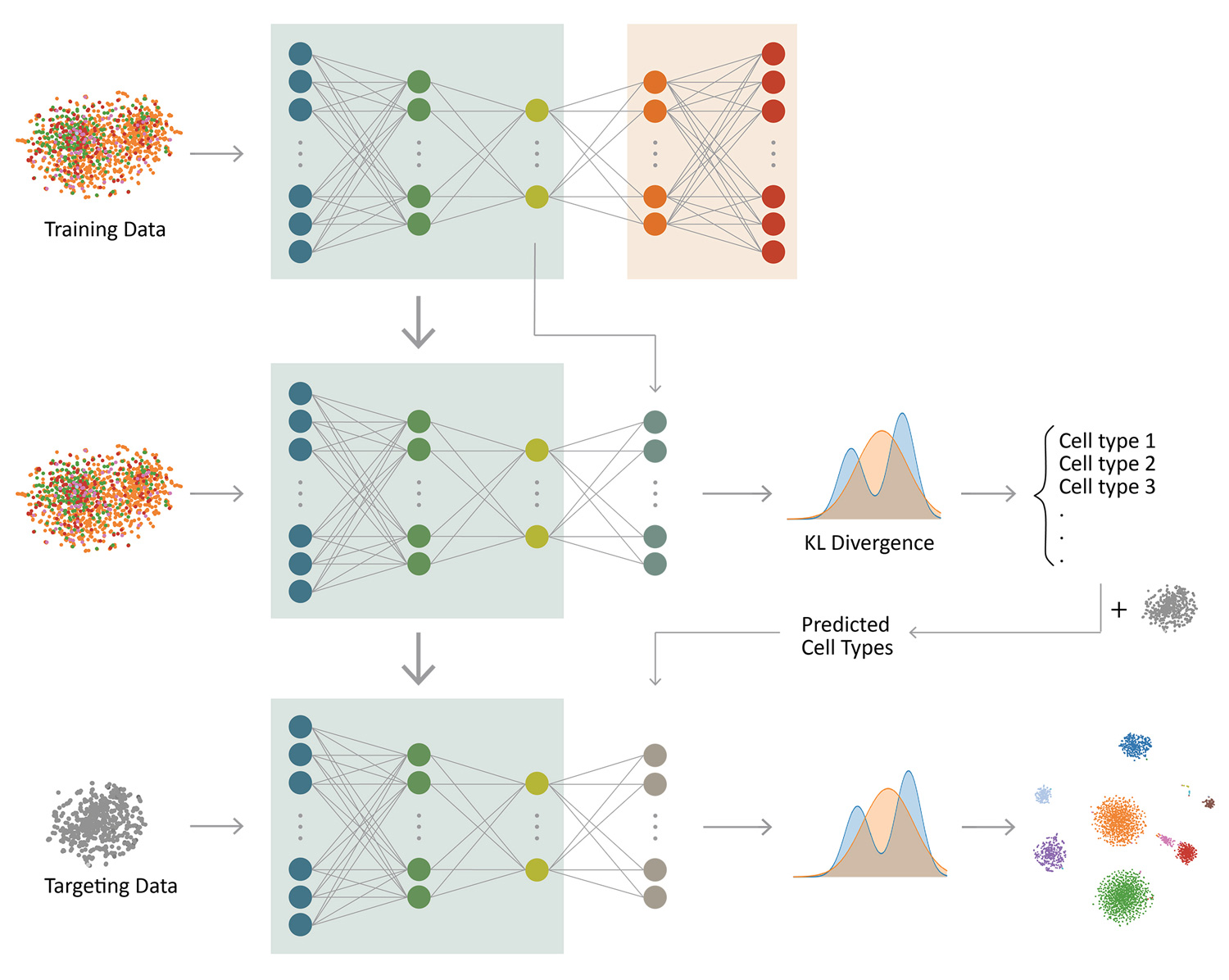
Clustering and cell type classification are important steps in single-cell RNA-seq (scRNA-seq) analysis. As more and more scRNA-seq data are becoming available, supervised cell type classification methods that utilize external well-annotated source data start to gain popularity over unsupervised clustering algorithms; however, the performance of existing supervised methods is highly dependent on source data quality and they often have limited accuracy to classify cell types that are missing in the source data. We developed ItClust to overcome these limitations, a transfer learning algorithm that borrows ideas from super- vised cell type classification algorithms, but also leverages information in target data to ensure sensitivity in classifying cells that are only present in the target data. Through extensive evaluations using data from different species and tissues generated with diverse scRNA-seq protocols, we show that ItClust considerably improves clustering and cell type classification accuracy over popular unsupervised clustering and supervised cell type classification algorithms.
