Jian Hu, Amelia Schroeder, Kyle Coleman, Chixiang Chen, Benjamin J Auerbach, Mingyao Li
Published in Computational and Structural Biotechnology Journal
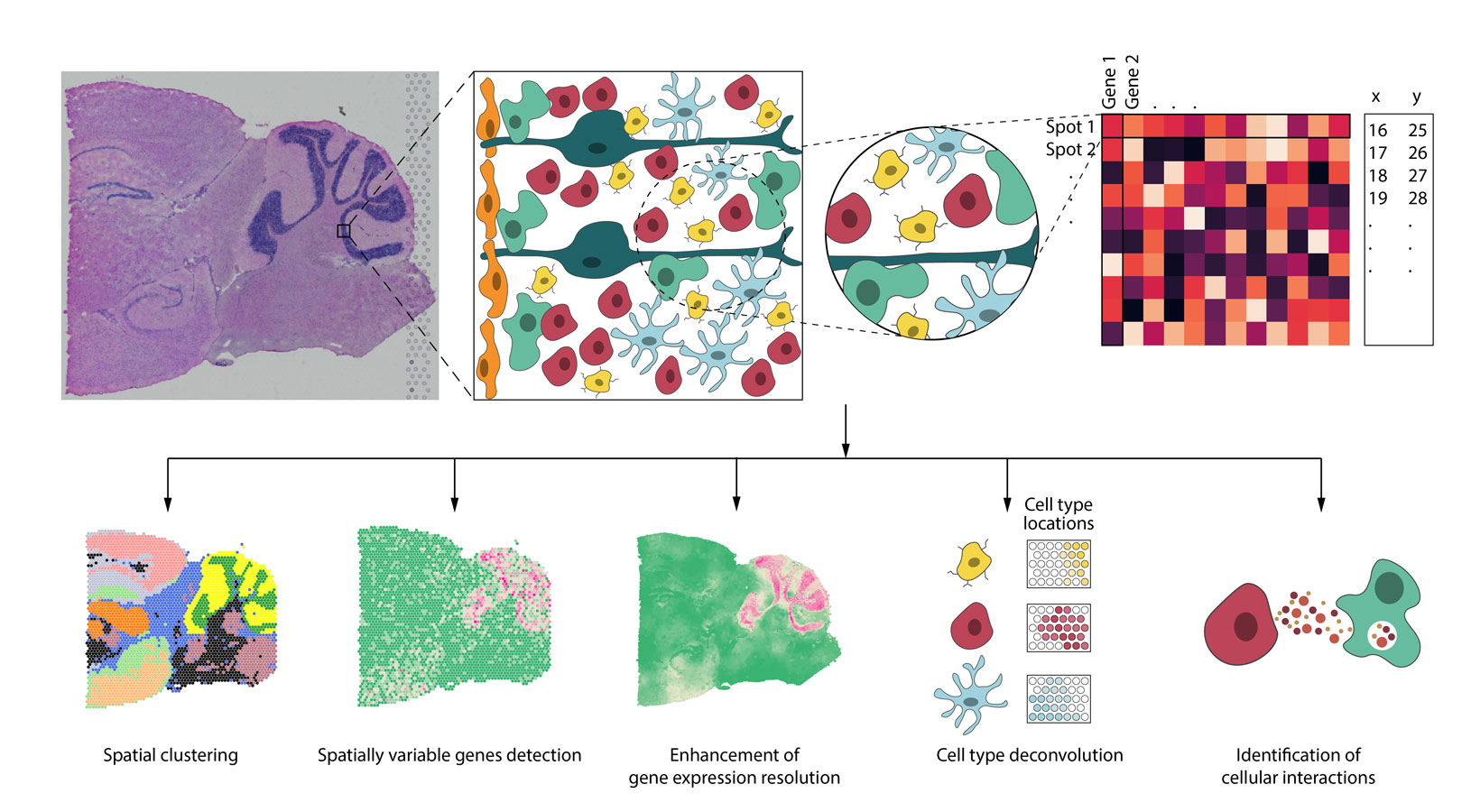
Spatial resolved transcriptomics (SRT) technologies have enabled scientists to get an integrated understanding of cells in their morphological context. Applications of these technologies in diverse tissues and diseases have transformed our views of transcriptional complexity. Most published studies utilized tools developed for single-cell RNA sequencing (scRNA-seq) for data analysis. However, SRT data exhibit different properties from scRNA-seq. To take full advantage of the added dimension on spatial location information in such data, new methods that are tailored for SRT are needed. Additionally, SRT data often have companion high-resolution histology information available. Incorporating histological features in gene expression analysis is an underexplored area. In this review, we will focus on the statistical and machine learning aspects for SRT data analysis and discuss how spatial location and histology information can be integrated with gene expression to advance our understanding of the transcriptional complexity. We also point out open problems and future research directions in this field.
